Note
Go to the end to download the full example code.
Decoupling template generation and alignment¶
When performing template-based alignment, one often uses the same subjects both to generate the template and to align to it. However, this can lead to overfitting and optimistic results (see Jeganathan et al.[1]). In this example, we illustrate how out-of-sample template generation can be performed seamlessly using fmralign.
To run this example, you must launch IPython via ipython --matplotlib in
a terminal, or use jupyter-notebook.
Retrieve the data¶
In this example we use the IBC dataset, which include a large number of different contrasts maps for 12 subjects. We download the images for subjects sub-01, sub-02 and sub-04 (or retrieve them if they were already downloaded).
from nilearn.maskers import NiftiMasker
from fmralign.fetch_example_data import fetch_ibc_subjects_contrasts
subjects = ["sub-01", "sub-02", "sub-04"]
files, df, mask = fetch_ibc_subjects_contrasts(subjects)
masker = NiftiMasker(mask_img=mask).fit()
[get_dataset_dir] Dataset found in /home/runner/nilearn_data/ibc
Generating an in-sample template¶
First, we generate templates using all subjects. This is the standard approach, and the fastest in terms of number of alignments to perform since only one template is generated for the whole group. We split the data into alignment data (task “archi_standard”) and test data (task “archi_spatial”).
from fmralign import GroupAlignment
from fmralign.embeddings.parcellation import get_labels
X_alignment = {
sub: masker.transform(
df[(df.subject == sub) & (df.task == "archi_standard")].path
)
for sub in subjects
}
X_test = {
sub: masker.transform(
df[(df.subject == sub) & (df.task == "archi_spatial")].path
)
for sub in subjects
}
# Use only the first image to speed up the computation of the labels
labels = get_labels(
masker.inverse_transform(X_alignment["sub-01"]),
n_pieces=150,
masker=masker,
)
population_algo = GroupAlignment("procrustes", labels=labels)
population_algo.fit(X_alignment, y="template")
/home/runner/work/fmralign/fmralign/fmralign/embeddings/parcellation.py:114: UserWarning: Converting masker to multi-masker for compatibility with Nilearn Parcellations class. This conversion does not affect the original masker. See https://github.com/nilearn/nilearn/issues/5926 for more details.
warnings.warn(
/home/runner/work/fmralign/fmralign/fmralign/embeddings/parcellation.py:134: UserWarning: Overriding provided-default estimator parameters with provided masker parameters :
Parameter mask_strategy :
Masker parameter background - overriding estimator parameter epi
Parameter smoothing_fwhm :
Masker parameter None - overriding estimator parameter 4.0
parcellation.fit(images_to_parcel)
/home/runner/work/fmralign/fmralign/.venv/lib/python3.12/site-packages/sklearn/cluster/_agglomerative.py:323: UserWarning: the number of connected components of the connectivity matrix is 35 > 1. Completing it to avoid stopping the tree early.
connectivity, n_connected_components = _fix_connectivity(
/home/runner/work/fmralign/fmralign/fmralign/alignment/utils.py:212: UserWarning:
Some parcels are more than 1000 voxels wide it can slow down alignment,especially optimal_transport :
parcel 1 : 1011 voxels
parcel 7 : 1315 voxels
parcel 30 : 1696 voxels
parcel 43 : 1846 voxels
warnings.warn(warning)
Generating an out-of-sample template¶
Now, we generate template alignments in a leave-one-subject-out fashion. For each subject, we generate a template using all other subjects, and align the left-out subject to this template. This ensures that template generation and alignments between the template and subjects are decoupled.
loso_algo = GroupAlignment("procrustes", labels=labels)
loso_algo.fit(X_alignment, y="leave_one_subject_out")
Aligning test data¶
Finally, we align the test data for each subject to both templates and compute the average aligned test data across subjects.
import numpy as np
aligned_in_sample = population_algo.transform(X_test)
aligned_out_of_sample = loso_algo.transform(X_test)
average_in_sample = np.mean(
[aligned_in_sample[sub] for sub in subjects], axis=0
)
average_out_of_sample = np.mean(
[aligned_out_of_sample[sub] for sub in subjects], axis=0
)
Comparing the correlations¶
We compare the average correlation of the transformed data across subjects for both in-sample and out-of-sample template generation strategies.
import matplotlib.pyplot as plt
from nilearn import plotting
from fmralign.metrics import score_voxelwise
score_in_sample = np.mean(
[
score_voxelwise(img, average_in_sample, loss="corr")
for img in aligned_in_sample.values()
],
axis=0,
)
score_in_sample_img = masker.inverse_transform(score_in_sample)
score_out_of_sample = np.mean(
[
score_voxelwise(img, average_out_of_sample, loss="corr")
for img in aligned_out_of_sample.values()
],
axis=0,
)
score_out_of_sample_img = masker.inverse_transform(score_out_of_sample)
fig, axes = plt.subplots(2, 1, figsize=(8, 12))
plotting.plot_stat_map(
score_in_sample_img,
display_mode="z",
cut_coords=[-5, -15],
vmax=1,
title="Inter-Subject Correlations (In-sample Template)",
axes=axes[0],
colorbar=True,
)
plotting.plot_stat_map(
score_out_of_sample_img,
display_mode="z",
cut_coords=[-5, -15],
vmax=1,
title="Inter-Subject Correlations (Out-of-sample Template)",
axes=axes[1],
colorbar=True,
)
plt.show()
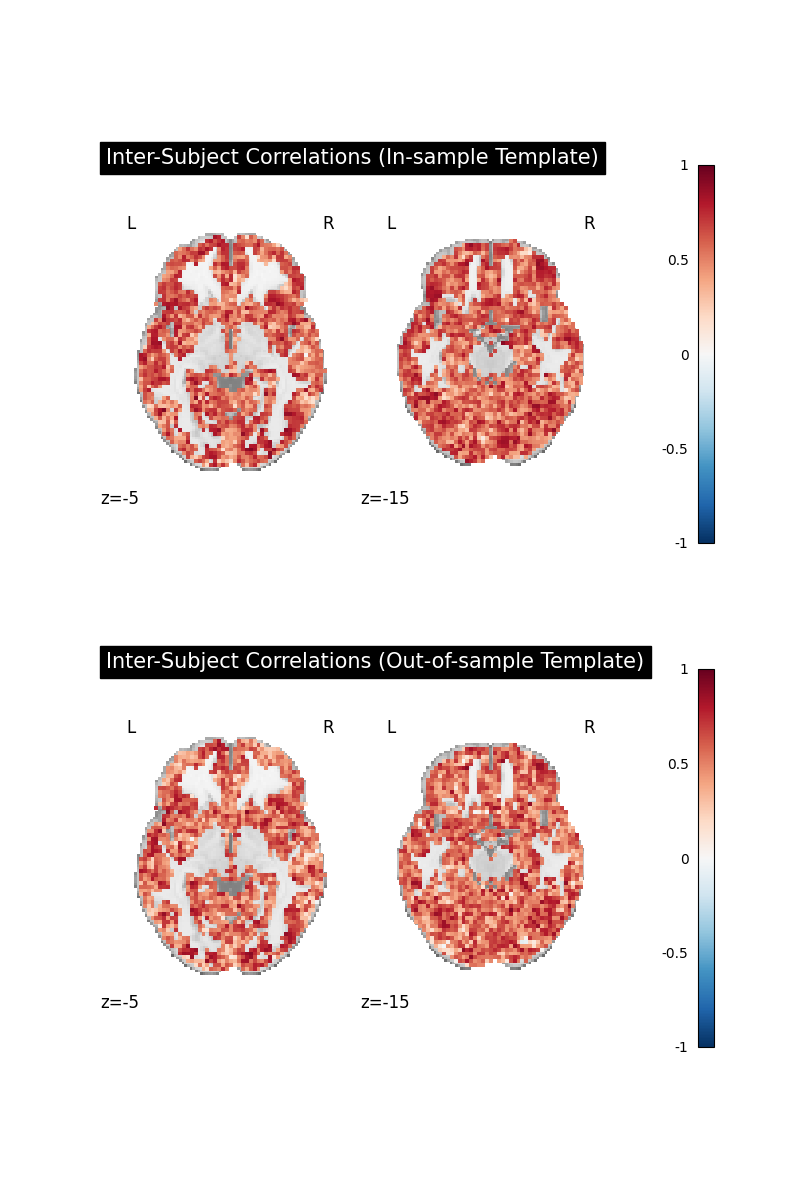
The direct comparisons of voxelwise inter-subject correlations show little difference
between in-sample and out-of-sample template generation strategies. That is good news,
as it indicates that in-sample template generation does not lead to a large bias
in this case.
To better visualize the differences, we can use nilearn’s
plot_img_comparison
plotting.plot_img_comparison(
score_in_sample_img,
score_out_of_sample_img,
masker=masker,
ref_label="In-sample Template",
src_label="Out-of-sample Template",
)
plt.show()
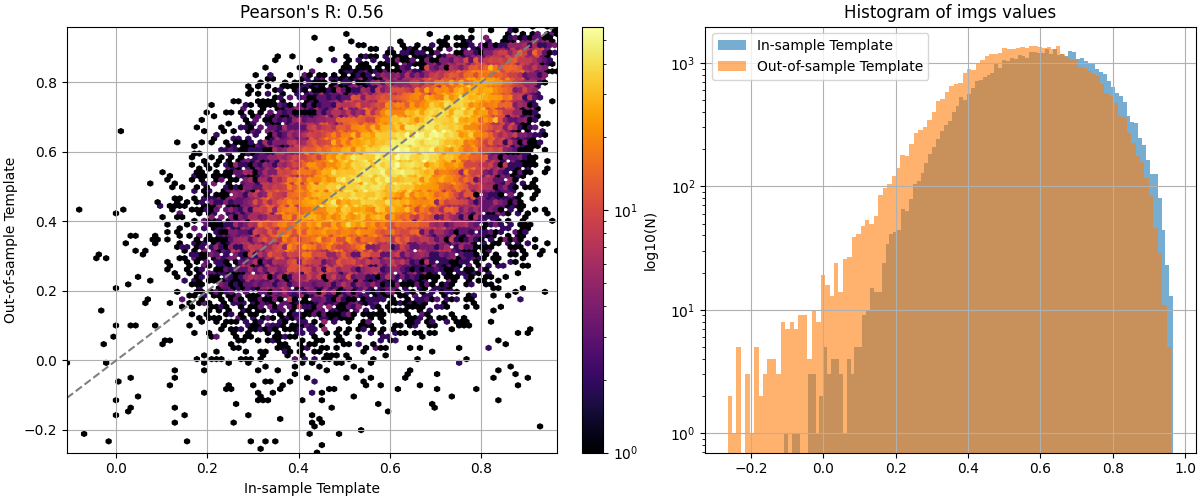
We are now able to see that in-sample template generation leads to slightly higher inter-subject correlations after alignment, indicating a small bias. To conclude, out-of-sample template generation avoids this bias at the cost of having to perform more alignments. As the number of subjects increases, the difference between both strategies narrows. However, when dealing with small datasets, out-of-sample template generation is recommended to avoid overly optimistic results.
References¶
Total running time of the script: (2 minutes 7.207 seconds)