Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
fmralign.embeddings.parcellation.get_labels¶
- fmralign.embeddings.parcellation.get_labels(imgs, masker, n_pieces=1, clustering='ward', smoothing_fwhm=5, verbose=0)[source]¶
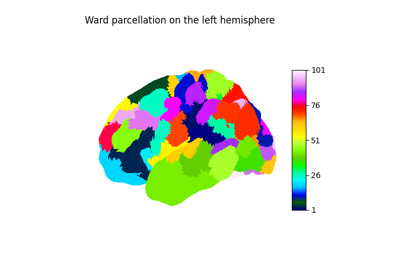
Generate an array of labels for each voxel in the data.
Use nilearn Parcellation class in our pipeline. It is used to find local regions of the brain in which alignment will be later applied. For alignment computational efficiency, regions should be of hundreds of voxels.
- Parameters:
- imgs: Niimgs
data to cluster
- n_pieces: int
number of different labels
- masker: a fitted instance of NiftiMasker or MultiNiftiMasker
Masker to be used on the data. For more information see: http://nilearn.github.io/manipulating_images/masker_objects.html
- clustering: string or 3D Niimg
If you aim for speed, choose k-means (and check kmeans_smoothing_fwhm parameter) If you want spatially connected and/or reproducible regions use ‘ward’ If you want balanced clusters (especially from timeseries) used ‘hierarchical_kmeans’ If 3D Niimg, image used as predefined clustering, n_pieces is ignored
- smoothing_fwhm: None or int
By default 5mm smoothing will be applied before kmeans clustering to have more compact clusters (but this will not change the data later). To disable this option, this parameter should be None.
- verbose: int, default=0
Verbosity level.
- Returns:
- labelslist of ints (len n_features)
Parcellation of features in clusters
Examples using fmralign.embeddings.parcellation.get_labels¶
Alignment methods benchmark (template-based ROI case)